3 Annotating land use
In this study we aim to quantify the response to fireworks across different bird communities, for which we use take-off habitat as a proxy. In this notebook we will classify land use, and a variety of factors that can serve as a proxy for the severity of fireworks disturbance, such as the distance to the nearest residential area for each of the PPI ‘pixels’.
The land use is based on the CORINE Land Cover dataset and specifically the 2018 version (CLC2018), which should be most relevant for the 2017-2018 fireworks event.
3.2 Converting the land use map
To start, we need to convert the land use map to the same 1) resolution, and 2) extent of the radar PPIs as we can then simply ‘overlay’ both rasters on top of each other and merge them. We load the PPIs and extract the CRS information contained in the proj4 strings.
ppi_hrw <- readRDS("data/processed/corrected_ppi_hrw.RDS")
ppi_dhl <- readRDS("data/processed/corrected_ppi_dhl.RDS")
ppi_proj4_hrw <- ppi_hrw$data@proj4string
ppi_proj4_dhl <- ppi_dhl$data@proj4stringAnd we load and prepare the land use map it’s all about. To aid the classification process, we also load the legend of land use classes contained in the entire CLC2018 dataset, otherwise the classes will remain anonymous numbers.
landuse <- raster("data/raw/landuse/clc2018_clc2018_v2018_20_raster100m/CLC2018_CLC2018_V2018_20.tif")
landuse_classes <- read.csv("data/raw/landuse/clc2018_clc2018_v2018_20_raster100m/Legend/CLC2018_CLC2018_V2018_20_QGIS.txt",
col.names = c("landuse_id", "r", "g", "b", "x", "landuse_class"), header = FALSE)[, c("landuse_id", "landuse_class"),]3.2.1 Cropping the land use map
As the CLC2018 dataset is so large it does not fit in memory at all in the steps below, so we have to crop the raster dataset for further processing. Even then, it still requires a beefy machine to process these files. First, we calculate a bounding box for the landuse raster based on the bounding boxes of the radar data.
padding <- 25000 # Padding in m to make sure we crop out of the land use map with a wide margin to compensate for edge-effects later
bbox_meters <- abs(ppi_dhl$data@bbox[[1]]) + padding # Assuming the PPI range of DHL and HRW are the same
bbox_hrw <- st_bbox(c(xmin = -bbox_meters, ymin = -bbox_meters, xmax = bbox_meters, ymax = bbox_meters), crs = ppi_proj4_hrw)
bbox_dhl <- st_bbox(c(xmin = -bbox_meters, ymin = -bbox_meters, xmax = bbox_meters, ymax = bbox_meters), crs = ppi_proj4_dhl)
bbox_hrw %>%
st_as_sfc() %>%
st_transform(crs(landuse)) %>%
st_bbox -> bbox_landuse_hrw
bbox_dhl %>%
st_as_sfc() %>%
st_transform(crs(landuse)) %>%
st_bbox -> bbox_landuse_dhlWe can now crop and plot the land use maps centered on the radar sites, with a meter padding surrounding the extent of the radar data.
ext_hrw <- extent(c(bbox_landuse_hrw[1], bbox_landuse_hrw[3], bbox_landuse_hrw[2], bbox_landuse_hrw[4]))
ext_dhl <- extent(c(bbox_landuse_dhl[1], bbox_landuse_dhl[3], bbox_landuse_dhl[2], bbox_landuse_dhl[4]))
landuse_crop_hrw <- crop(landuse, ext_hrw)## Warning in showSRID(uprojargs, format = "PROJ", multiline = "NO", prefer_proj
## = prefer_proj): Discarded datum Unknown based on GRS80 ellipsoid in Proj4
## definition
landuse_crop_dhl <- crop(landuse, ext_dhl)## Warning in showSRID(uprojargs, format = "PROJ", multiline = "NO", prefer_proj
## = prefer_proj): Discarded datum Unknown based on GRS80 ellipsoid in Proj4
## definition
sea_id <- match('Sea and ocean', landuse_classes$landuse_class)
landuse_crop_hrw[is.na(landuse_crop_hrw)] <- landuse_classes[sea_id, "landuse_id"] # Convert NAs to the land use class of the sea
landuse_crop_dhl[is.na(landuse_crop_dhl)] <- landuse_classes[sea_id, "landuse_id"]And plot the result:
par(pty = "s", mfrow = c(1, 2))
image(landuse_crop_hrw, main = "Herwijnen")
image(landuse_crop_dhl, main = "Den Helder")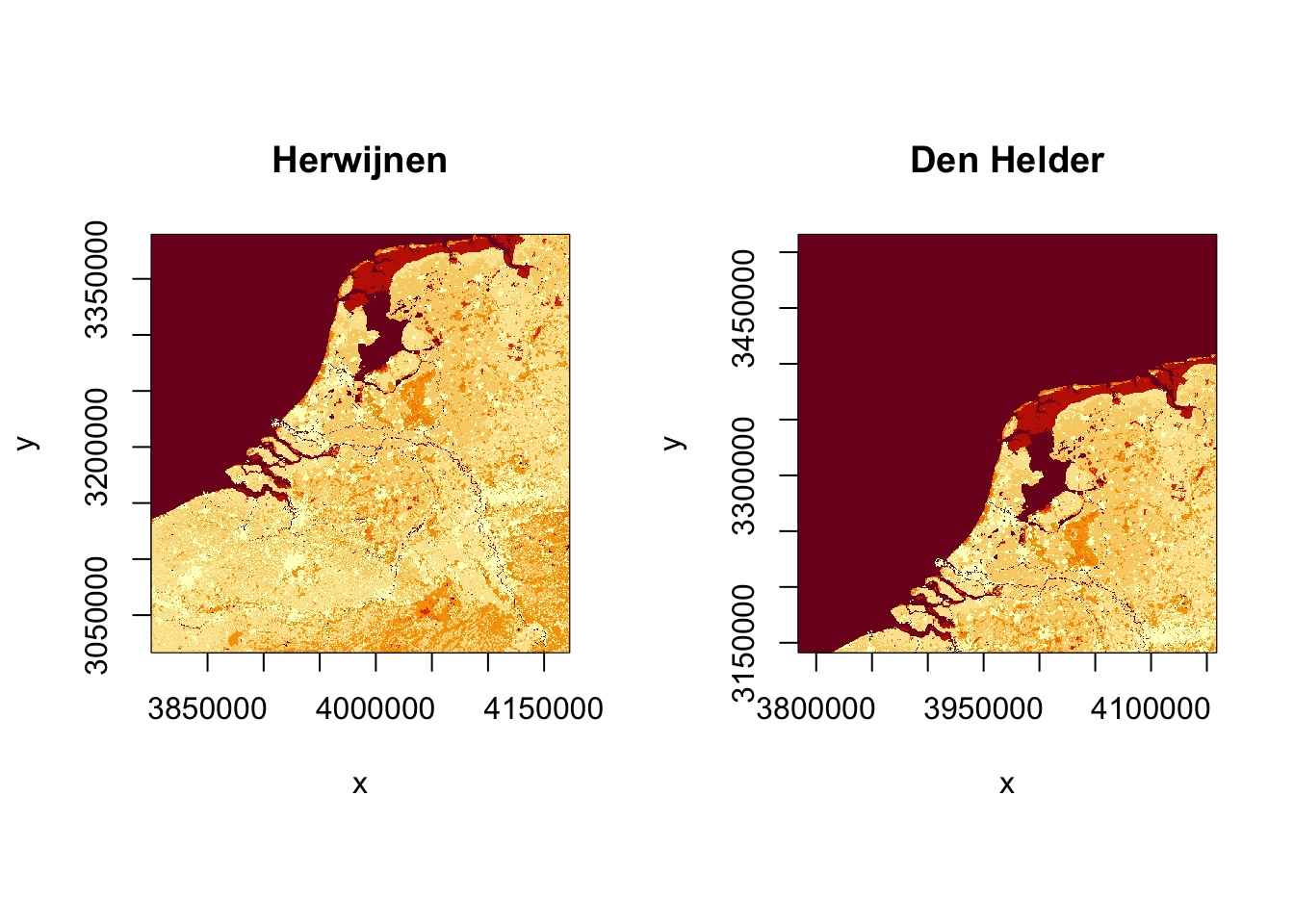
3.2.2 Reprojecting the land use map
Now that the land use map is cropped, we can start the reprojection to the CRS of the radar PPI. As we’re dealing with categorical data, we set method = "ngb" to use nearest neighbour interpolation. Despite using interpolation, this reprojection does not change the resolution from the base of the CLC2018 dataset to that of the PPI, so we’ll have to do that next.
landuse_hrw_reprojected <- projectRaster(landuse_crop_hrw, crs = ppi_proj4_hrw, method = "ngb")
landuse_dhl_reprojected <- projectRaster(landuse_crop_dhl, crs = ppi_proj4_dhl, method = "ngb")
levels(landuse_hrw_reprojected) <- levels(landuse_crop_hrw)
levels(landuse_dhl_reprojected) <- levels(landuse_crop_dhl)If the reprojection went successful, the CRS of the reprojected land use map and the radar PPI should be the same.
compareCRS(ppi_hrw$data@proj4string, landuse_hrw_reprojected@crs)
compareCRS(ppi_dhl$data@proj4string, landuse_dhl_reprojected@crs)## [1] TRUE
## [1] TRUEApparently that is the case.
3.2.3 Reclassifying land use types to functional classes
The CLC2018 dataset contains a total of 44 land use classes. For our purpose, we reduce these 44 to 5 classes with more biologically relevant groupings, specifically:
- Urban area
- Agricultural area
- Semi-open area
- Forests
- Wetlands
- Water bodies
The following table is used to convert/reclassify the classes contained within the CLC2018 dataset, with the original land use classes under landuse_class and how they will be reclassified under landuse_target. We also indicate whether these areas are inhabited (inhabited = 1), or uninhabited (inhabited = 0). In The Netherlands inhabited areas are classified as “Discontinuous urban fabric” in the CLC2018 dataset, all other areas we set to uninhabited.
landuse_classes %<>%
mutate(landuse_target = case_when(
landuse_id >= 100 & landuse_id < 200 ~ "Urban area",
landuse_id >= 200 & landuse_id <= 213 ~ "Agricultural area",
landuse_id >= 221 & landuse_id <= 223 ~ "Semi-open area",
landuse_id >= 231 & landuse_id <= 243 ~ "Agricultural area",
landuse_id >= 244 & landuse_id < 300 ~ "Forests",
landuse_id >= 300 & landuse_id <= 313 ~ "Forests",
landuse_id >= 321 & landuse_id <= 335 ~ "Semi-open area",
landuse_id >= 400 & landuse_id < 500 ~ "Wetlands",
landuse_id >= 500 & landuse_id < 999 ~ "Water bodies",
landuse_id == 999 ~ "NODATA")) %>%
mutate(landuse_target_id = case_when(
landuse_target == "Urban area" ~ 1,
landuse_target == "Agricultural area" ~ 2,
landuse_target == "Semi-open area" ~ 3,
landuse_target == "Forests" ~ 4,
landuse_target == "Wetlands" ~ 5,
landuse_target == "Water bodies" ~ 6,
landuse_target == "NODATA" ~ 9
))
landuse_classes$inhabited <- 0
landuse_classes$inhabited[landuse_classes$landuse_class == "Discontinuous urban fabric"] <- 1
landuse_classes| landuse_id | landuse_class | landuse_target | landuse_target_id | inhabited |
|---|---|---|---|---|
| 111 | Continuous urban fabric | Urban area | 1 | 0 |
| 112 | Discontinuous urban fabric | Urban area | 1 | 1 |
| 121 | Industrial or commercial units | Urban area | 1 | 0 |
| 122 | Road and rail networks and associated land | Urban area | 1 | 0 |
| 123 | Port areas | Urban area | 1 | 0 |
| 124 | Airports | Urban area | 1 | 0 |
| 131 | Mineral extraction sites | Urban area | 1 | 0 |
| 132 | Dump sites | Urban area | 1 | 0 |
| 133 | Construction sites | Urban area | 1 | 0 |
| 141 | Green urban areas | Urban area | 1 | 0 |
| 142 | Sport and leisure facilities | Urban area | 1 | 0 |
| 211 | Non-irrigated arable land | Agricultural area | 2 | 0 |
| 212 | Permanently irrigated land | Agricultural area | 2 | 0 |
| 213 | Rice fields | Agricultural area | 2 | 0 |
| 221 | Vineyards | Semi-open area | 3 | 0 |
| 222 | Fruit trees and berry plantations | Semi-open area | 3 | 0 |
| 223 | Olive groves | Semi-open area | 3 | 0 |
| 231 | Pastures | Agricultural area | 2 | 0 |
| 241 | Annual crops associated with permanent crops | Agricultural area | 2 | 0 |
| 242 | Complex cultivation patterns | Agricultural area | 2 | 0 |
| 243 | Land principally occupied by agriculture with significant areas of natural vegetation | Agricultural area | 2 | 0 |
| 244 | Agro-forestry areas | Forests | 4 | 0 |
| 311 | Broad-leaved forest | Forests | 4 | 0 |
| 312 | Coniferous forest | Forests | 4 | 0 |
| 313 | Mixed forest | Forests | 4 | 0 |
| 321 | Natural grasslands | Semi-open area | 3 | 0 |
| 322 | Moors and heathland | Semi-open area | 3 | 0 |
| 323 | Sclerophyllous vegetation | Semi-open area | 3 | 0 |
| 324 | Transitional woodland-shrub | Semi-open area | 3 | 0 |
| 331 | Beaches dunes sands | Semi-open area | 3 | 0 |
| 332 | Bare rocks | Semi-open area | 3 | 0 |
| 333 | Sparsely vegetated areas | Semi-open area | 3 | 0 |
| 334 | Burnt areas | Semi-open area | 3 | 0 |
| 335 | Glaciers and perpetual snow | Semi-open area | 3 | 0 |
| 411 | Inland marshes | Wetlands | 5 | 0 |
| 412 | Peat bogs | Wetlands | 5 | 0 |
| 421 | Salt marshes | Wetlands | 5 | 0 |
| 422 | Salines | Wetlands | 5 | 0 |
| 423 | Intertidal flats | Wetlands | 5 | 0 |
| 511 | Water courses | Water bodies | 6 | 0 |
| 512 | Water bodies | Water bodies | 6 | 0 |
| 521 | Coastal lagoons | Water bodies | 6 | 0 |
| 522 | Estuaries | Water bodies | 6 | 0 |
| 523 | Sea and ocean | Water bodies | 6 | 0 |
| 999 | NODATA | NODATA | 9 | 0 |
With a sort-of ‘raster attribute table’ in place, we can now reclassify the detailed landuse classes to the broader categories listed above.
landuse_dhl_reclassified <- ratify(reclassify(landuse_dhl_reprojected, cbind(landuse_classes$landuse_id, landuse_classes$landuse_target_id)), count = TRUE)
landuse_hrw_reclassified <- ratify(reclassify(landuse_hrw_reprojected, cbind(landuse_classes$landuse_id, landuse_classes$landuse_target_id)), count = TRUE)
writeRaster(landuse_dhl_reclassified, "data/processed/landuse/landuse_dhl_reclassified.tif", overwrite = TRUE)
writeRaster(landuse_hrw_reclassified, "data/processed/landuse/landuse_hrw_reclassified.tif", overwrite = TRUE)This leaves us with the following count of ~100x100m cells per land use category
cellcounts <- cbind(levels(landuse_hrw_reclassified)[[1]]["COUNT"], levels(landuse_dhl_reclassified)[[1]]["COUNT"])
colnames(cellcounts) <- c("Herwijnen", "Den Helder")
rownames(cellcounts) <- unique(landuse_classes$landuse_target)[-7]
cellcounts| Herwijnen | Den Helder | |
|---|---|---|
| Urban area | 1860678 | 809932 |
| Agricultural area | 6256242 | 3384907 |
| Semi-open area | 194271 | 133709 |
| Forests | 1437047 | 498510 |
| Wetlands | 346616 | 364301 |
| Water bodies | 3851726 | 8890353 |
3.2.4 Resampling the land use map to a lower resolution
The cellsize of the PPIs is 500x500 meters, but the land use map is much more finely detailed (~100x100m), so we need to resample the latter to derive a land use map at a 500x500 meter resolution as well.
As the resolution of the PPIs is so much higher than that of the land use map, we need to resample the latter to a lower resolution. Instead of classifying a single pixel in the PPI as belonging to a single land use class, we will do so using land use proportions. We therefore calculate the proportions belonging to each of the land use classes within an area of cells roughly the size of the PPI pixels. Subsequently we resample this to match 1:1 with the PPI pixels and store the land use proportions for each of the land use classes in the PPIs. This is done in the calculate_land_use_proportion() function below.
calculate_land_use_proportion <- function(raster, reference_raster, overwrite = FALSE) {
values <- c(sort(unique(getValues(raster))))
# classes: multidimensional logical array for the classes contained within the land use map
# 1 if a land use class is present on that position, 0 if not
classes <- layerize(raster, filename = paste("data/processed/landuse/layerize/", substitute(raster), sep = ""),
format = "raster", bylayer = TRUE, classes = values, overwrite = overwrite)
# factor: nr of cells in both horizontal and vertical direction to aggregate
factor <- round(dim(raster)[1:2] / dim(reference_raster)[1:2])
# agg: aggregated version of classes (aggregation factor defined by factor) containing mean coverage by a class in a given area
# 1 corresponds with full coverage, 0 with no coverage of that class within the pixel at all
agg <- aggregate(classes, factor, na.rm = TRUE, fun = mean)
# x: the agg and ppi pixels almost overlap exactly, but there is a teeny tiny difference which we can
# iron out by resampling once more.
x <- resample(agg, reference_raster)
return(x)
}We can now calculate the proportions and will save these to some raster files for potential inspection in GIS software.
landuse_hrw <- calculate_land_use_proportion(landuse_hrw_reclassified, as(ppi_hrw$data, "RasterLayer"), overwrite = TRUE)
landuse_dhl <- calculate_land_use_proportion(landuse_dhl_reclassified, as(ppi_dhl$data, "RasterLayer"), overwrite = TRUE)
names(landuse_hrw) <- c("urban", "agricultural", "semiopen", "forests", "wetlands", "waterbodies")
names(landuse_dhl) <- c("urban", "agricultural", "semiopen", "forests", "wetlands", "waterbodies")
writeRaster(landuse_hrw, "data/processed/landuse/landuse_hrw.tif", overwrite = TRUE)
writeRaster(landuse_dhl, "data/processed/landuse/landuse_dhl.tif", overwrite = TRUE)By now the resampled land use raster should be very similar to the PPI raster, with the exception of — of course — the values contained within.
compareRaster(landuse_hrw, as(ppi_hrw$data, "RasterLayer"), extent = TRUE, rowcol = TRUE, crs = TRUE, res = TRUE, orig = TRUE)
compareRaster(landuse_dhl, as(ppi_dhl$data, "RasterLayer"), extent = TRUE, rowcol = TRUE, crs = TRUE, res = TRUE, orig = TRUE)## [1] TRUE
## [1] TRUEOk, let’s save a copy of what we have so far.
3.3 Adding land use classifications to the PPIs
With the land use rasters overlapping exactly with the PPIs, we can simply extract the values of the resampled land use rasters and add these as additional parameters to the PPIs.
landuse_hrw <- readRDS("data/processed/landuse/landuse_hrw.RDS")
landuse_dhl <- readRDS("data/processed/landuse/landuse_dhl.RDS")
values_hrw <- rasterToPoints(landuse_hrw, spatial = TRUE)
values_dhl <- rasterToPoints(landuse_dhl, spatial = TRUE)
ppi_hrw$data@data <- cbind(ppi_hrw$data@data, values_hrw@data)
ppi_dhl$data@data <- cbind(ppi_dhl$data@data, values_dhl@data)3.4 Calculate distance to nearest urban area
We can use the distance to the nearest inhabited urban area as a proxy for disturbance. To calculate this, we reclassify the raster to cells containing inhabited urban area and everything else. For every cell on the raster that is not a cell we have just classified as inhabited urban area, we will calculate the distance (in meters) to the nearest cell classified as inhabited urban area. We classify a PPI cell as uninhabited if the proportion of its constituent cells (at a finer resolution) that is inhabited is > 0.95.
dhl_inhabited <- ratify(reclassify(landuse_dhl_reprojected, cbind(landuse_classes$landuse_id, landuse_classes$inhabited)), count = TRUE)
hrw_inhabited <- ratify(reclassify(landuse_hrw_reprojected, cbind(landuse_classes$landuse_id, landuse_classes$inhabited)), count = TRUE)
dhl_inhabited <- calculate_land_use_proportion(dhl_inhabited, as(ppi_dhl$data, "RasterLayer"), overwrite = TRUE)
hrw_inhabited <- calculate_land_use_proportion(hrw_inhabited, as(ppi_hrw$data, "RasterLayer"), overwrite = TRUE)
names(dhl_inhabited) <- c("uninhabited", "inhabited")
names(hrw_inhabited) <- c("uninhabited", "inhabited")
dist_dhl <- dhl_inhabited
dist_dhl[dist_dhl$uninhabited > 0.95] <- NA
dist_dhl <- distance(dist_dhl$inhabited)
dist_hrw <- hrw_inhabited
dist_hrw[dist_hrw$uninhabited > 0.95] <- NA
dist_hrw <- distance(dist_hrw$inhabited)
writeRaster(dist_hrw, "data/processed/landuse/dist_urban_hrw.tif", overwrite = TRUE)
writeRaster(dist_dhl, "data/processed/landuse/dist_urban_dhl.tif", overwrite = TRUE)And we add these values to the PPIs again.
values_dist_hrw <- rasterToPoints(dist_hrw, spatial = TRUE)
values_dist_dhl <- rasterToPoints(dist_dhl, spatial = TRUE)
ppi_hrw$data@data$dist_urban <- values_dist_hrw@data$layer
ppi_dhl$data@data$dist_urban <- values_dist_dhl@data$layer3.5 Add population density
Another proxy for disturbance to explore is simply the number of humans living in a certain area. The Dutch Central Bureau of Statistics (CBS) annually publishes a dataset containing the number of inhabitants organized in 500x500m grid cells. We will now add this to the PPIs as well.
cbs_maps <- st_read("data/raw/population-density/2019-CBS_VK500_2018_v1/CBS_VK500_2018_v1.shp")## Reading layer `CBS_VK500_2018_v1' from data source
## `/Users/barthoekstra/Development/fireworks/data/raw/population-density/2019-CBS_VK500_2018_v1/CBS_VK500_2018_v1.shp'
## using driver `ESRI Shapefile'
## Simple feature collection with 151108 features and 31 fields
## Geometry type: POLYGON
## Dimension: XY
## Bounding box: xmin: 13000 ymin: 306500 xmax: 278500 ymax: 619500
## Projected CRS: Amersfoort / RD New
cbs_hrw <- st_transform(cbs_maps, ppi_hrw$data@proj4string)
cbs_dhl <- st_transform(cbs_maps, ppi_dhl$data@proj4string)
# Template for the rasterization following the standard 500x500m resolution of the CBS grid
template_hrw <- st_as_stars(st_bbox(cbs_hrw["INWONER"]), values = NA_real_, dx = 500, dy = 500)
template_dhl <- st_as_stars(st_bbox(cbs_dhl["INWONER"]), values = NA_real_, dx = 500, dy = 500)
# Now rasterize
pop_density_rasterized_hrw <- as(st_rasterize(cbs_hrw["INWONER"], template = template_hrw), "Raster")
pop_density_rasterized_dhl <- as(st_rasterize(cbs_dhl["INWONER"], template = template_dhl), "Raster")
# Set negative or NA raster values to 0
pop_density_rasterized_hrw[pop_density_rasterized_hrw < 0] <- 0
pop_density_rasterized_dhl[pop_density_rasterized_dhl < 0] <- 0
# Now aggregate by summing up values in cells to 'cover' the values that fit in a PPI pixel
factor_hrw <- round(dim(pop_density_rasterized_hrw)[1:2] / dim(as(ppi_hrw$data, "RasterLayer"))[1:2])
factor_dhl <- round(dim(pop_density_rasterized_dhl)[1:2] / dim(as(ppi_dhl$data, "RasterLayer"))[1:2])
agg_hrw <- aggregate(pop_density_rasterized_hrw, factor_hrw, na.rm = TRUE, fun = sum)## Warning in .local(x, ...): all fact(s) were 1, nothing to aggregate
agg_dhl <- aggregate(pop_density_rasterized_dhl, factor_dhl, na.rm = TRUE, fun = sum)## Warning in .local(x, ...): all fact(s) were 1, nothing to aggregate
# Resample to make the PPI and CBS population grids line up 1:1
pop_density_rasterized_hrw <- resample(agg_hrw, as(ppi_hrw$data, "RasterLayer"))
pop_density_rasterized_dhl <- resample(agg_dhl, as(ppi_dhl$data, "RasterLayer"))Once again, verify if the PPI pixels match up exactly with the CBS population grids
compareRaster(pop_density_rasterized_hrw, as(ppi_hrw$data, "RasterLayer"), extent = TRUE, rowcol = TRUE, crs = TRUE, res = TRUE, orig = TRUE)
compareRaster(pop_density_rasterized_dhl, as(ppi_dhl$data, "RasterLayer"), extent = TRUE, rowcol = TRUE, crs = TRUE, res = TRUE, orig = TRUE)## [1] TRUE
## [1] TRUENow as a final step we will calculate the total population within the neighborhood surrounding the PPI pixels, to get a more representative measure of disturbance potential in the surrounding area.
weights <- matrix(1, nrow = 3, ncol = 3)
pop_hrw <- focal(pop_density_rasterized_hrw, w = weights, fun = sum, na.rm = TRUE)
pop_dhl <- focal(pop_density_rasterized_dhl, w = weights, fun = sum, na.rm = TRUE)
pop_hrw[is.na(pop_hrw)] <- 0
pop_dhl[is.na(pop_dhl)] <- 0And add it to the PPIs.
values_pop_hrw <- rasterToPoints(pop_hrw, spatial = TRUE)
values_pop_dhl <- rasterToPoints(pop_dhl, spatial = TRUE)
ppi_hrw$data@data$human_pop <- values_pop_hrw@data$layer
ppi_dhl$data@data$human_pop <- values_pop_dhl@data$layer3.6 Add land area logical
Over land we have created adequately filtered PPIs in previous steps, but over at the North Sea there can still be dynamic clutter remaining caused by ships and wind parks. To eventually quantify the number of birds affected by fireworks in our scan, we should be able to filter VIR only for areas above land, where we are quite sure clutter has been filtered sufficiently. The approach will be similar to how we have added population density in the previous step.
# We get data from the CBS, which can be done dynamically through the command below, but because the geodata service occasionally hangs, we stored the
# data locally as well in data/raw/municipalities/municipalities.json
# mcp <- st_read("https://geodata.nationaalgeoregister.nl/wijkenbuurten2020/ows?request=GetFeature&service=WFS&version=1.1.0&typeName=gemeenten2020&outputFormat=json")
mcp <- st_read("data/raw/municipalities/municipalities.json")## Reading layer `municipalities' from data source
## `/Users/barthoekstra/Development/fireworks/data/raw/municipalities/municipalities.json'
## using driver `GeoJSON'
## Simple feature collection with 438 features and 35 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: 10425.16 ymin: 306846.2 xmax: 278026.1 ymax: 621876.3
## Projected CRS: Amersfoort / RD New
mcp_hrw <- st_transform(mcp, ppi_hrw$data@proj4string)
mcp_dhl <- st_transform(mcp, ppi_dhl$data@proj4string)
# Template for the rasterization following the standard 500x500m resolution of the CBS grid
template_hrw <- st_as_stars(st_bbox(mcp_hrw["gemeentenaam"]), values = NA_real_, dx = 500, dy = 500)
template_dhl <- st_as_stars(st_bbox(mcp_dhl["gemeentenaam"]), values = NA_real_, dx = 500, dy = 500)
# Now rasterize
mcp_rasterized_hrw <- as(st_rasterize(mcp_hrw["gemeentenaam"], template = template_hrw), "Raster")
mcp_rasterized_dhl <- as(st_rasterize(mcp_dhl["gemeentenaam"], template = template_dhl), "Raster")
# Set NA to 0, non-NA to 1
mcp_rasterized_hrw[!is.na(mcp_rasterized_hrw)] <- 1
mcp_rasterized_hrw[is.na(mcp_rasterized_hrw)] <- 0
mcp_rasterized_dhl[!is.na(mcp_rasterized_dhl)] <- 1
mcp_rasterized_dhl[is.na(mcp_rasterized_dhl)] <- 0
# Resample to PPI grid
mcp_rasterized_hrw <- resample(mcp_rasterized_hrw, as(ppi_hrw$data, "RasterLayer"))
mcp_rasterized_dhl <- resample(mcp_rasterized_dhl, as(ppi_dhl$data, "RasterLayer"))
# Set NA to 0 again
mcp_rasterized_hrw[is.na(mcp_rasterized_hrw)] <- 0
mcp_rasterized_dhl[is.na(mcp_rasterized_dhl)] <- 0Once again, verify if the PPI pixels match up exactly with the gridded land area.
compareRaster(mcp_rasterized_hrw, as(ppi_hrw$data, "RasterLayer"), extent = TRUE, rowcol = TRUE, crs = TRUE, res = TRUE, orig = TRUE)
compareRaster(mcp_rasterized_dhl, as(ppi_dhl$data, "RasterLayer"), extent = TRUE, rowcol = TRUE, crs = TRUE, res = TRUE, orig = TRUE)## [1] TRUE
## [1] TRUEWe can now add the land area to the PPIs again.
values_land_hrw <- rasterToPoints(mcp_rasterized_hrw, spatial = TRUE)
values_land_dhl <- rasterToPoints(mcp_rasterized_dhl, spatial = TRUE)
ppi_hrw$data@data$land <- values_land_hrw@data$layer
ppi_dhl$data@data$land <- values_land_dhl@data$layerAnd finally we save these PPIs again.
saveRDS(ppi_hrw, file = "data/processed/corrected_ppi_hrw_lu.RDS")
saveRDS(ppi_dhl, file = "data/processed/corrected_ppi_dhl_lu.RDS")3.6.1 Plotting human parameters on an interactive map
We have now generated proxies for fireworks ‘severity’ for later modelling stages. Let’s plot them on an interactive map again for reference. Unfortunately, due to the way leaflet stores maps it produces, we cannot plot them here using K-M knitting to generate this document. Run these chunks interactively and they should work fine.
3.6.1.1 Interactive map of Herwijnen
human_parameters <- c("urban", "agricultural", "semiopen", "forests", "wetlands", "waterbodies", "dist_urban", "human_pop", "land")
mapview(ppi_hrw$data[, , human_parameters], alpha.regions = 0.6, col.regions = inferno, maxpixels=2000000,
na.color = "#00000000", map.types = c("CartoDB.Positron", "CartoDB.DarkMatter", "Esri.WorldImagery"),
layer.name = human_parameters)